R Exercises – 31-40 – Data Frame Manipulations
1. Working with the ‘mtcars’ dataset
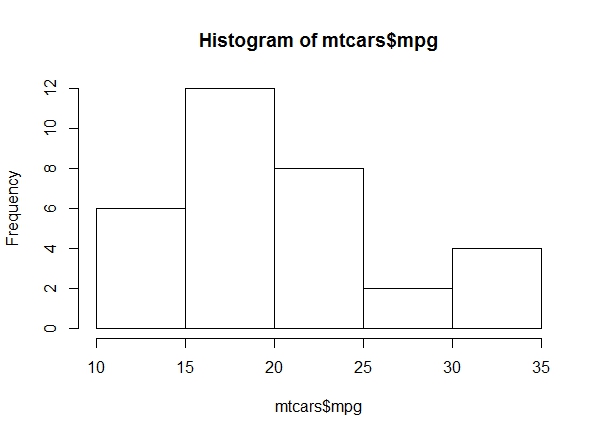
a. Get a histogram of the ‘mpg’ values of ‘mtcars’. Which bin contains the most observations?
b. Are there more automatic (0) or manual (1) transmission-type cars in the dataset? Hint: ‘mtcars’ has 32 observations.
c. Get a scatter plot of ‘hp’ vs ‘weight’.
hist(mtcars$mpg)Bin 15-20
b.
sum(mtcars$am == 1)
sum(mtcars$am == 0)There are more automatic (0) cars.
c.
plot(mtcars$wt, mtcars$hp, col = "red", pch = 3,
bty = "l", xlab = "Car Weight in 1K lbs", ylab = "Horsepower" )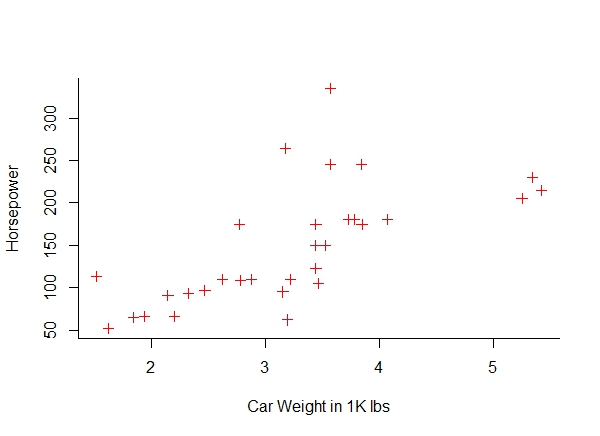
2. Working with the ‘iris’ dataset
a. Get all rows of Species ‘versicolor’ in a new data frame. Call this data frame: ‘iris.vers’
#expected result Sepal.Length Sepal.Width Petal.Length Petal.Width Species 51 7.0 3.2 4.7 1.4 versicolor 52 6.4 3.2 4.5 1.5 versicolor 53 6.9 3.1 4.9 1.5 versicolor 54 5.5 2.3 4.0 1.3 versicolor 55 6.5 2.8 4.6 1.5 versicolor 56 5.7 2.8 4.5 1.3 versicolor 57 6.3 3.3 4.7 1.6 versicolor 58 4.9 2.4 3.3 1.0 versicolor 59 6.6 2.9 4.6 1.3 versicolor 60 5.2 2.7 3.9 1.4 versicolor
b. Get a vector called ‘sepal.dif’ with the difference between ‘Sepal.Length’ and ‘Sepal.Width’ of ‘versicolor’ plants.
#expected result [1] 3.8 3.2 3.8 3.2 3.7 2.9 3.0 2.5 3.7 2.5 3.0 2.9 3.8 3.2 2.7 3.6 2.6 3.1 4.0 [20] 3.1 2.7 3.3 3.8 3.3 3.5 3.6 4.0 3.7 3.1 3.1 3.1 3.1 3.1 3.3 2.4 2.6 3.6 4.0 [39] 2.6 3.0 2.9 3.1 3.2 2.7 2.9 2.7 2.8 3.3 2.6 2.9
c. Update (add) ‘iris.vers’ with the new column ‘sepal.dif’.
#expected result Sepal.Length Sepal.Width Petal.Length Petal.Width Species sepal.dif 51 7.0 3.2 4.7 1.4 versicolor 3.8 52 6.4 3.2 4.5 1.5 versicolor 3.2 53 6.9 3.1 4.9 1.5 versicolor 3.8 54 5.5 2.3 4.0 1.3 versicolor 3.2 55 6.5 2.8 4.6 1.5 versicolor 3.7 56 5.7 2.8 4.5 1.3 versicolor 2.9
iris.vers = subset(iris, Species == "versicolor"); iris.versb.
sepal.dif = iris.vers$Sepal.Length - iris.vers$Sepal.Width; sepal.difc.
iris.vers = data.frame(iris.vers, sepal.dif); head(iris.vers) 3. Classes of Variables (‘mtcars’)
a. Check the class of each variable in ‘mtcars’.
#expected result
mpg cyl disp hp drat wt qsec vs
"numeric" "numeric" "numeric" "numeric" "numeric" "numeric" "numeric" "numeric"
am gear carb
"numeric" "numeric" "numeric"
b. Change ‘am’, ‘cyl’ and ‘vs’ to integer and store the new dataset as ‘newmtc’.
#expected result
mpg as.integer.cyl. disp hp drat
"numeric" "integer" "numeric" "numeric" "numeric"
wt qsec as.integer.vs. as.integer.am. gear
"numeric" "numeric" "integer" "integer" "numeric"
carb
"numeric"
c. Round the ‘newmtc’ data frame to one digit.
#expected result
mpg as.integer.cyl. disp hp drat wt qsec as.integer.vs. as.integer.am. gear carb
1 21.0 6 160.0 110 3.9 2.6 16.5 0 1 4 4
2 21.0 6 160.0 110 3.9 2.9 17.0 0 1 4 4
3 22.8 4 108.0 93 3.8 2.3 18.6 1 1 4 1
4 21.4 6 258.0 110 3.1 3.2 19.4 1 0 3 1
5 18.7 8 360.0 175 3.1 3.4 17.0 0 0 3 2
6 18.1 6 225.0 105 2.8 3.5 20.2 1 0 3 1
7 14.3 8 360.0 245 3.2 3.6 15.8 0 0 3 4
8 24.4 4 146.7 62 3.7 3.2 20.0 1 0 4 2
9 22.8 4 140.8 95 3.9 3.1 22.9 1 0 4 2
10 19.2 6 167.6 123 3.9 3.4 18.3 1 0 4 4
head(mtcars)
sapply(mtcars, class)b.
attach(mtcars)
newmtc = data.frame(mpg, as.integer(cyl), disp, hp, drat, wt, qsec,
as.integer(vs), as.integer(am), gear, carb)
sapply(newmtc, class)c.
round(newmtc, 1) 4. Advanced filtering of ‘iris’
a. Use ‘dplyr’ to filter for all data of Species ‘virginica’ with a ‘Sepal.Width’ of greater than 3.5.
#expected result Sepal.Length Sepal.Width Petal.Length Petal.Width Species 1 7.2 3.6 6.1 2.5 virginica 2 7.7 3.8 6.7 2.2 virginica 3 7.9 3.8 6.4 2.0 virginica
b. How would you use R Base to get a data frame of all data of Species ‘virginica’ with a ‘Sepal.Width’ of greater than 3.5, but without the last column Species in the data frame?
#expected result
Sepal.Length Sepal.Width Petal.Length Petal.Width
110 7.2 3.6 6.1 2.5
118 7.7 3.8 6.7 2.2
132 7.9 3.8 6.4 2.0
c. Get the row IDs of the rows matching the two filtering criteria provided above.
#expected result [1] "110" "118" "132"
library(dplyr)
filter(iris, Sepal.Width > 3.5, Species =="virginica")b.
iris[iris$Species == "virginica" & iris$Sepal.Width > 3.5, 1:4]c.
row.names(iris[iris$Species == "virginica" & iris$Sepal.Width > 3.5, 1:4]) 5. Manipulating data frames at column level (dataset = ‘iris’)
a. Repeat each value of ‘Sepal.Length’ two times and repeat the whole sequence two times as well.
#expected result [1] 5.1 5.1 4.9 4.9 4.7 4.7 4.6 4.6 5.0 5.0 5.4 5.4 4.6 4.6 5.0 5.0 4.4 4.4 4.9 4.9 5.4 5.4 4.8 [24] 4.8 4.8 4.8 4.3 4.3 5.8 5.8 5.7 5.7 5.4 5.4 5.1 5.1 5.7 5.7 5.1 5.1 5.4 5.4 5.1 5.1 4.6 4.6 [47] 5.1 5.1 4.8 4.8 5.0 5.0 5.0 5.0 5.2 5.2 5.2 5.2 4.7 4.7 4.8 4.8 5.4 5.4 5.2 5.2 5.5 5.5 4.9 [70] 4.9 5.0 5.0 5.5 5.5 4.9 4.9 4.4 4.4 5.1 5.1 5.0 5.0 4.5 4.5 4.4 4.4 5.0 5.0 5.1 5.1 4.8 4.8 [93] 5.1 5.1 4.6 4.6 5.3 5.3 5.0 5.0 7.0 7.0 6.4 6.4 6.9 6.9 5.5 5.5 6.5 6.5 5.7 5.7 6.3 6.3 4.9 [116] 4.9 6.6 6.6 5.2 5.2 5.0 5.0 5.9 5.9 6.0 6.0 6.1 6.1 5.6 5.6 6.7 6.7 5.6 5.6 5.8 5.8 6.2 6.2 [139] 5.6 5.6 5.9 5.9 6.1 6.1 6.3 6.3 6.1 6.1 6.4 6.4 6.6 6.6 6.8 6.8 6.7 6.7 6.0 6.0 5.7 5.7 5.5 [162] 5.5 5.5 5.5 5.8 5.8 6.0 6.0 5.4 5.4 6.0 6.0 6.7 6.7 6.3 6.3 5.6 5.6 5.5 5.5 5.5 5.5 6.1 6.1 [185] 5.8 5.8 5.0 5.0 5.6 5.6 5.7 5.7 5.7 5.7 6.2 6.2 5.1 5.1 5.7 5.7 6.3 6.3 5.8 5.8 7.1 7.1 6.3 [208] 6.3 6.5 6.5 7.6 7.6 4.9 4.9 7.3 7.3 6.7 6.7 7.2 7.2 6.5 6.5 6.4 6.4 6.8 6.8 5.7 5.7 5.8 5.8 [231] 6.4 6.4 6.5 6.5 7.7 7.7 7.7 7.7 6.0 6.0 6.9 6.9 5.6 5.6 7.7 7.7 6.3 6.3 6.7 6.7 7.2 7.2 6.2 [254] 6.2 6.1 6.1 6.4 6.4 7.2 7.2 7.4 7.4 7.9 7.9 6.4 6.4 6.3 6.3 6.1 6.1 7.7 7.7 6.3 6.3 6.4 6.4 [277] 6.0 6.0 6.9 6.9 6.7 6.7 6.9 6.9 5.8 5.8 6.8 6.8 6.7 6.7 6.7 6.7 6.3 6.3 6.5 6.5 6.2 6.2 5.9 [300] 5.9 5.1 5.1 4.9 4.9 4.7 4.7 4.6 4.6 5.0 5.0 5.4 5.4 4.6 4.6 5.0 5.0 4.4 4.4 4.9 4.9 5.4 5.4 [323] 4.8 4.8 4.8 4.8 4.3 4.3 5.8 5.8 5.7 5.7 5.4 5.4 5.1 5.1 5.7 5.7 5.1 5.1 5.4 5.4 5.1 5.1 4.6 [346] 4.6 5.1 5.1 4.8 4.8 5.0 5.0 5.0 5.0 5.2 5.2 5.2 5.2 4.7 4.7 4.8 4.8 5.4 5.4 5.2 5.2 5.5 5.5 [369] 4.9 4.9 5.0 5.0 5.5 5.5 4.9 4.9 4.4 4.4 5.1 5.1 5.0 5.0 4.5 4.5 4.4 4.4 5.0 5.0 5.1 5.1 4.8 [392] 4.8 5.1 5.1 4.6 4.6 5.3 5.3 5.0 5.0 7.0 7.0 6.4 6.4 6.9 6.9 5.5 5.5 6.5 6.5 5.7 5.7 6.3 6.3 [415] 4.9 4.9 6.6 6.6 5.2 5.2 5.0 5.0 5.9 5.9 6.0 6.0 6.1 6.1 5.6 5.6 6.7 6.7 5.6 5.6 5.8 5.8 6.2 [438] 6.2 5.6 5.6 5.9 5.9 6.1 6.1 6.3 6.3 6.1 6.1 6.4 6.4 6.6 6.6 6.8 6.8 6.7 6.7 6.0 6.0 5.7 5.7 [461] 5.5 5.5 5.5 5.5 5.8 5.8 6.0 6.0 5.4 5.4 6.0 6.0 6.7 6.7 6.3 6.3 5.6 5.6 5.5 5.5 5.5 5.5 6.1 [484] 6.1 5.8 5.8 5.0 5.0 5.6 5.6 5.7 5.7 5.7 5.7 6.2 6.2 5.1 5.1 5.7 5.7 6.3 6.3 5.8 5.8 7.1 7.1 [507] 6.3 6.3 6.5 6.5 7.6 7.6 4.9 4.9 7.3 7.3 6.7 6.7 7.2 7.2 6.5 6.5 6.4 6.4 6.8 6.8 5.7 5.7 5.8 [530] 5.8 6.4 6.4 6.5 6.5 7.7 7.7 7.7 7.7 6.0 6.0 6.9 6.9 5.6 5.6 7.7 7.7 6.3 6.3 6.7 6.7 7.2 7.2 [553] 6.2 6.2 6.1 6.1 6.4 6.4 7.2 7.2 7.4 7.4 7.9 7.9 6.4 6.4 6.3 6.3 6.1 6.1 7.7 7.7 6.3 6.3 6.4 [576] 6.4 6.0 6.0 6.9 6.9 6.7 6.7 6.9 6.9 5.8 5.8 6.8 6.8 6.7 6.7 6.7 6.7 6.3 6.3 6.5 6.5 6.2 6.2 [599] 5.9 5.9
b. Get a new object which contains only the odd values of ‘Sepal.Length’.
#expected result [1] 5.1 4.7 5.0 4.6 4.4 5.4 4.8 5.8 5.4 5.7 5.4 4.6 4.8 5.0 5.2 4.8 5.2 4.9 5.5 4.4 5.0 4.4 5.1 [24] 5.1 5.3 7.0 6.9 6.5 6.3 6.6 5.0 6.0 5.6 5.6 6.2 5.9 6.3 6.4 6.8 6.0 5.5 5.8 5.4 6.7 5.6 5.5 [47] 5.8 5.6 5.7 5.1 6.3 7.1 6.5 4.9 6.7 6.5 6.8 5.8 6.5 7.7 6.9 7.7 6.7 6.2 6.4 7.4 6.4 6.1 6.3 [70] 6.0 6.7 5.8 6.7 6.3 6.2
c. Get a new object which repeats each value from the new vector of exercise b.
#expected result [1] 5.1 5.1 4.7 4.7 5.0 5.0 4.6 4.6 4.4 4.4 5.4 5.4 4.8 4.8 5.8 5.8 5.4 5.4 5.7 5.7 5.4 5.4 4.6 [24] 4.6 4.8 4.8 5.0 5.0 5.2 5.2 4.8 4.8 5.2 5.2 4.9 4.9 5.5 5.5 4.4 4.4 5.0 5.0 4.4 4.4 5.1 5.1 [47] 5.1 5.1 5.3 5.3 7.0 7.0 6.9 6.9 6.5 6.5 6.3 6.3 6.6 6.6 5.0 5.0 6.0 6.0 5.6 5.6 5.6 5.6 6.2 [70] 6.2 5.9 5.9 6.3 6.3 6.4 6.4 6.8 6.8 6.0 6.0 5.5 5.5 5.8 5.8 5.4 5.4 6.7 6.7 5.6 5.6 5.5 5.5 [93] 5.8 5.8 5.6 5.6 5.7 5.7 5.1 5.1 6.3 6.3 7.1 7.1 6.5 6.5 4.9 4.9 6.7 6.7 6.5 6.5 6.8 6.8 5.8 [116] 5.8 6.5 6.5 7.7 7.7 6.9 6.9 7.7 7.7 6.7 6.7 6.2 6.2 6.4 6.4 7.4 7.4 6.4 6.4 6.1 6.1 6.3 6.3 [139] 6.0 6.0 6.7 6.7 5.8 5.8 6.7 6.7 6.3 6.3 6.2 6.2
d. Replace the ‘Sepal.Length’ column of ‘iris’ with the new ‘Sepal.Length’ from exercise c. Check if the replacement worked.
#expected result Sepal.Length Sepal.Width Petal.Length Petal.Width Species 1 5.1 3.5 1.4 0.2 setosa 2 5.1 3.0 1.4 0.2 setosa 3 4.7 3.2 1.3 0.2 setosa 4 4.7 3.1 1.5 0.2 setosa 5 5.0 3.6 1.4 0.2 setosa 6 5.0 3.9 1.7 0.4 setosa
rep(iris$Sepal.Length, each = 2, times = 2)b.
sep.lengthodd = iris[c(T,F),1]c.
newsep.length = rep(sep.lengthodd, each = 2)d.
iris$Sepal.Length = newsep.length; head(iris) 6. Big Data Example with ‘diamonds’ (package: ‘ggplot2’)
a. Get familiar with the dataset ‘diamonds’ from ‘ggplot2’.
b. Attach the dataset.
c. Get a subset from the diamonds dataset with all the rows that have a clarity of ‘SI2’ and a depth of at least 70. Call the subset ‘diam.sd’ and display it in the same line of code.
#expected result # A tibble: 6 x 10 carat cut color clarity depth table price x y z <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl> 1 1.50 Fair I SI2 70.1 58 4328 6.96 6.85 4.84 2 2.00 Fair F SI2 70.2 57 15351 7.63 7.59 5.34 3 0.70 Fair D SI2 71.6 55 1696 5.47 5.28 3.85 4 0.70 Fair E SI2 70.6 56 1828 5.45 5.35 3.81 5 1.00 Fair G SI2 70.2 58 2326 6.00 5.73 4.13 6 0.96 Fair G SI2 72.2 56 2438 6.01 5.81 4.28
d. Which index positions have a clarity of ‘SI2’ and a depth of at least 70? (hint: ‘row.names’)
#expected result [1] "1" "2" "3" "4" "5" "6"
e. Store the index positions as an integer object.
#expected result [1] 1 2 3 4 5 6
library(ggplot2)
? diamonds
summary(diamonds)b.
attach(diamonds)c.
diam.sd = subset(diamonds, clarity == "SI2" & depth >= 70); diam.sdd.
row.names(diam.sd)e.
index.pos = as.integer(row.names(diam.sd)); index.pos 7. Getting counts of filtered datasets (‘ggplot2’ : ‘diamonds’)
a. How many observations of diamonds have a cut of ‘ideal’ and have less than 0.21 carat?
b. How many observations of diamonds have a combined ‘x’ + ‘y’ + ‘z’ dimension greater than 40?
c. How many observations of diamonds have either a price above 10.000 USD or a depth of at least 70?
sum(cut == "Ideal" & carat < 0.21)'diamonds' is attached to environment
b.
sum ((x + y + z) > 40)c.
sum(price > 10000 | depth >= 70) 8. Filtering based on row and column ID (‘ggplot2’: ‘diamonds’)
a. Get a data frame with observations ’67’ and ‘982’ of variables color and y.
#expected result # A tibble: 2 x 2 color y <ord> <dbl> 1 I 4.42 2 F 5.76
b. Get a data frame with the full info on observations ‘453’, ‘792’ and ‘10489’.
#expected result # A tibble: 3 x 10 carat cut color clarity depth table price x y z <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl> 1 0.71 Ideal I VVS2 60.2 56 2817 5.84 5.89 3.53 2 0.70 Very Good D SI1 62.5 55 2862 5.67 5.72 3.56 3 1.01 Ideal F SI2 60.0 57 4796 6.55 6.51 3.92
c. Get the first 10 rows of the dataset ‘diamonds’ with the variables ‘x’, ‘y’, ‘z’.
#expected result
# A tibble: 10 x 3
x y z
<dbl> <dbl> <dbl>
1 3.95 3.98 2.43
2 3.89 3.84 2.31
3 4.05 4.07 2.31
4 4.20 4.23 2.63
5 4.34 4.35 2.75
6 3.94 3.96 2.48
7 3.95 3.98 2.47
8 4.07 4.11 2.53
9 3.87 3.78 2.49
10 4.00 4.05 2.39
d. Get the first six values of ‘diamonds’ of the variable ‘y’ as a simple vector.
#expected result
# A tibble: 6 x 1
y
<dbl>
1 3.98
2 3.84
3 4.07
4 4.23
5 4.35
6 3.96
diamonds[c(67,982), c(3,9)]b.
diamonds[c(453, 792, 10489), ]c.
head(diamonds[ , c(8,9,10)],10)d.
head(diamonds[,"y"])different output format: 'vector'
9. Ordering data (‘ggplot2’: ‘diamonds’)
a. Create the object ‘newdiam’ which is a subset of the first 1000 rows of ‘diamonds’.
#expected result # A tibble: 1,000 x 10 carat cut color clarity depth table price x y z <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl> 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31 3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31 4 0.29 Premium I VS2 62.4 58 334 4.20 4.23 2.63 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75 6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48 7 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47
b. Order ‘newdiam’ according to price, starting with the lowest. Hint: ‘dplyr’, ‘arrange’ is a useful function for that.
#expected result # A tibble: 6 x 10 carat cut color clarity depth table price x y z <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl> 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31 3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31 4 0.29 Premium I VS2 62.4 58 334 4.20 4.23 2.63 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75 6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
c. Order ‘newdiam’ according to price, starting with the highest.
#expected result # A tibble: 6 x 10 carat cut color clarity depth table price x y z <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl> 1 1.12 Premium J SI2 60.6 59 2898 6.68 6.61 4.03 2 0.60 Very Good D VVS2 60.6 57 2897 5.48 5.51 3.33 3 0.76 Premium E SI1 61.1 58 2897 5.91 5.85 3.59 4 0.54 Ideal D VVS2 61.4 52 2897 5.30 5.34 3.26 5 0.72 Ideal E SI1 62.5 55 2897 5.69 5.74 3.57 6 0.72 Good F VS1 59.4 61 2897 5.82 5.89 3.48
d. Order ‘newdiam’ like in exercise c, but ties should be arranged with increasing depth.
#expected result # A tibble: 6 x 10 carat cut color clarity depth table price x y z <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl> 1 1.12 Premium J SI2 60.6 59 2898 6.68 6.61 4.03 2 0.72 Good F VS1 59.4 61 2897 5.82 5.89 3.48 3 0.60 Very Good D VVS2 60.6 57 2897 5.48 5.51 3.33 4 0.76 Premium E SI1 61.1 58 2897 5.91 5.85 3.59 5 0.54 Ideal D VVS2 61.4 52 2897 5.30 5.34 3.26 6 0.74 Premium D VS2 61.8 58 2897 5.81 5.77 3.58
newdiam = diamonds[1:1000,]b.
library(dplyr)
head(arrange(newdiam, price))c.
head(arrange(newdiam, desc(price)))d.
head(arrange(newdiam, desc(price), depth)) 10. Sampling big data frames (‘ggplot2’: ‘diamonds’)
a. Use ‘dplyr’, ‘sample_n’ to get the object ‘diam750’ which contains 750 randomly sampled observations of ‘diamonds’. I use seed nr. 56 for reproduction.
#expected result # A tibble: 750 x 10 carat cut color clarity depth table price x y z <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl> 1 1.50 Ideal I VS1 61.3 57 9900 7.32 7.35 4.50 2 0.33 Ideal E VVS1 60.2 57 1052 4.50 4.47 2.70 3 1.02 Premium F VS2 62.4 59 6652 6.40 6.45 4.01 4 0.41 Premium F VS1 62.6 58 1076 4.75 4.74 2.97 5 1.08 Premium F VS2 62.1 59 7637 6.59 6.54 4.08 6 1.01 Premium I SI2 62.6 55 3818 6.43 6.38 4.01 7 1.05 Very Good G SI1 59.7 58 5019 6.60 6.66 3.96 8 1.00 Ideal F SI2 61.2 57 4452 6.48 6.39 3.94 9 0.55 Premium H SI2 59.8 59 1011 5.33 5.30 3.18 10 2.51 Fair H SI2 64.7 57 18308 8.44 8.50 5.48 # ... with 740 more rows
b. Get a summary of the new data frame.
#expected result
carat cut color clarity depth table
Min. :0.2300 Fair : 25 D: 73 SI1 :171 Min. :56.70 Min. :53.00
1st Qu.:0.4000 Good : 70 E:144 VS2 :154 1st Qu.:61.00 1st Qu.:56.00
Median :0.7050 Very Good:182 F:128 SI2 :141 Median :61.80 Median :57.00
Mean :0.8192 Premium :187 G:169 VS1 :120 Mean :61.77 Mean :57.51
3rd Qu.:1.0500 Ideal :286 H:127 VVS2 : 72 3rd Qu.:62.60 3rd Qu.:59.00
Max. :4.1300 I: 65 VVS1 : 56 Max. :69.70 Max. :67.00
J: 44 (Other): 36
price x y z
Min. : 361.0 Min. : 3.910 Min. :3.960 Min. :0.000
1st Qu.: 982.2 1st Qu.: 4.700 1st Qu.:4.710 1st Qu.:2.913
Median : 2385.5 Median : 5.700 Median :5.705 Median :3.535
Mean : 4066.9 Mean : 5.765 Mean :5.766 Mean :3.556
3rd Qu.: 5400.8 3rd Qu.: 6.548 3rd Qu.:6.555 3rd Qu.:4.050
Max. :18804.0 Max. :10.000 Max. :9.850 Max. :6.430
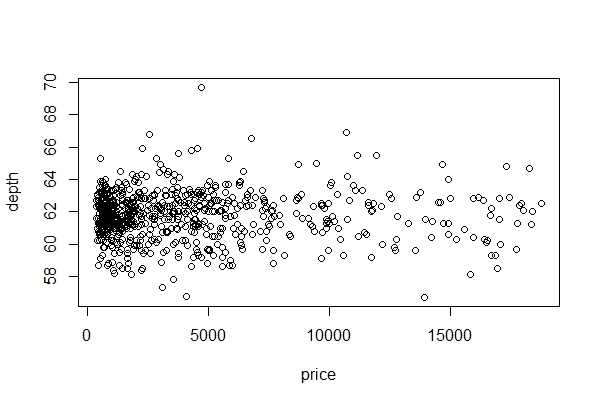
c. Plot a scatter plot of price vs depth. Use R Base plot, and the function ‘with’ (less code).
library(dplyr)
set.seed(56)
diam750 = sample_n(diamonds, 750)b.
summary(diam750)c.
with(diam750, plot(price, depth))